Metamodel Visualization#
When evaluating meta models, it can be useful to determine their fit of the training data, graphically.
OpenMDAO has a visualization package to view the training data and meta models generated from it.
This page explains how to use view_mm in the command line.
The metamodel viewer allows a user the ability of reducing a high dimensional input space down to three dimensions to enable the user to determine the fit of a meta model to the given training data.
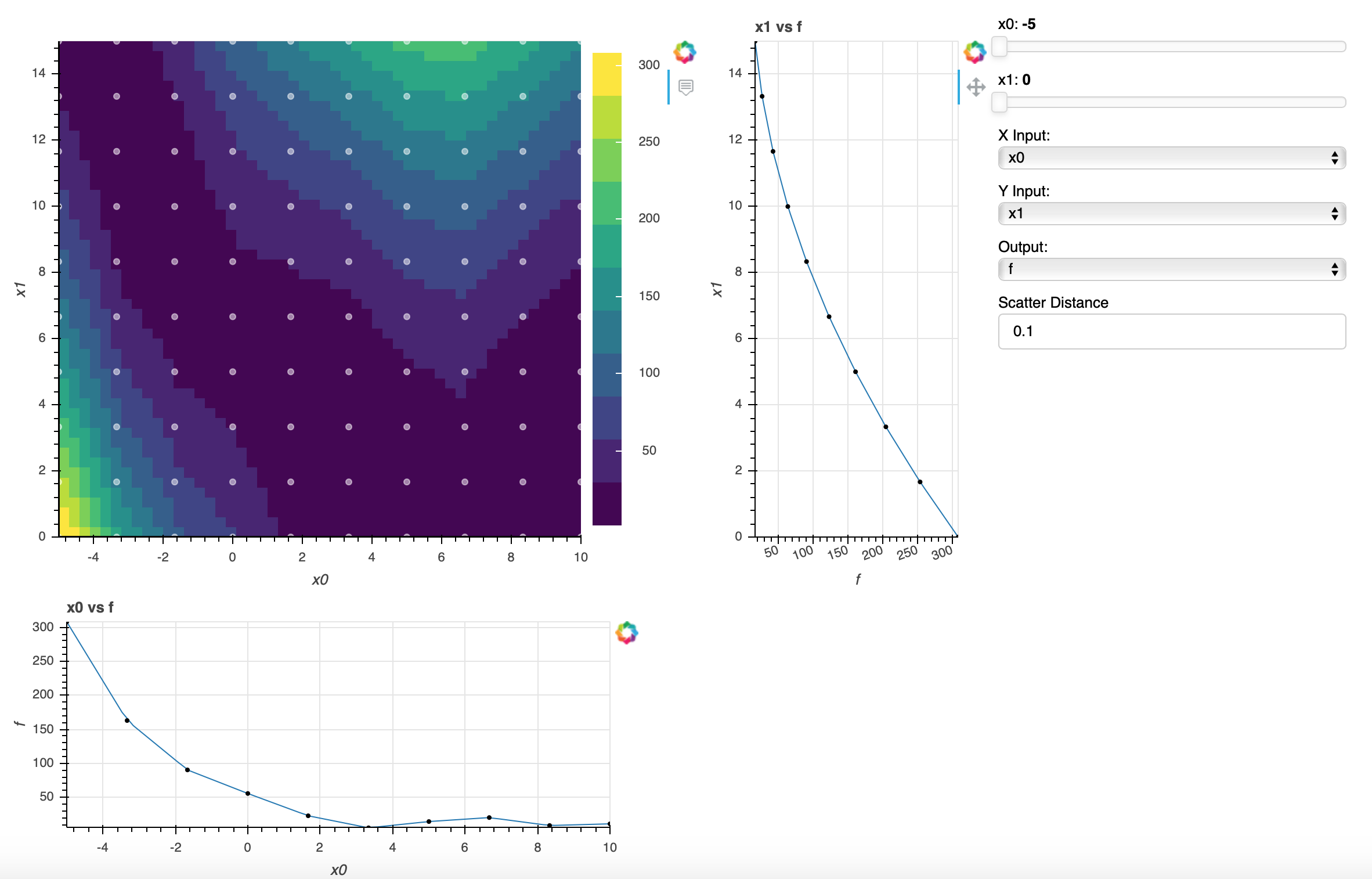
Running the Visualizer#
Note
This tool is accessible through the OpenMDAO command line tools which can be found om-command.
Running openmdao view_mm structured_meta_model_example.py will open the metamodel generated
from the script in the browser and generate a metamodel viewer like the one below. The user can adjust
sliders to make slices in the graph, change X and Y inputs, and change the scatter distance value to
fine tune the distance a point can be from the model line.
To recreate the viewer above, copy the first script given below and paste it into a file named structured_meta_model_example.py. Next, run openmdao view_mm structured_meta_model_example.py in the command line.
Structured MetaModel Example Script#
import numpy as np
import openmdao.api as om
num_train = 10
x0_min, x0_max = -5.0, 10.0
x1_min, x1_max = 0.0, 15.0
train_x0 = np.linspace(x0_min, x0_max, num_train)
train_x1 = np.linspace(x1_min, x1_max, num_train)
t_data = np.array([[308.12909601, 253.61567418, 204.6578079, 161.25549718, 123.40874201, 91.1175424, 64.38189835, 43.20180985, 27.5772769, 17.50829952],
[162.89542418, 123.20470795, 89.06954726, 60.48994214, 37.46589257, 19.99739855, 8.08446009, 1.72707719, 0.92524984, 5.67897804,],
[ 90.2866907, 63.02637433, 41.32161352, 25.17240826, 14.57875856, 9.54066442, 10.05812583, 16.13114279, 27.75971531, 44.94384339,],
[ 55.60211264, 38.37989042, 26.71322375, 20.60211264, 20.04655709, 25.04655709, 35.60211264, 51.71322375, 73.37989042, 100.60211264],
[ 22.81724065, 13.24080685, 9.2199286, 10.75460591, 17.84483877, 30.49062719, 48.69197117, 72.4488707, 101.76132579, 136.62933643],
[ 5.11168719, 0.78873608, 2.02134053, 8.80950054, 21.1532161, 39.05248721, 62.50731389, 91.51769611, 126.0836339, 166.20512723],
[ 14.3413983, 12.87962416, 16.97340558, 26.62274256, 41.82763509, 62.58808317, 88.90408682, 120.77564601, 158.20276077, 201.18543108],
[ 20.18431209, 19.1914092, 23.75406186, 33.87227009, 49.54603386, 70.77535319, 97.56022808, 129.90065853, 167.79664453, 211.24818608],
[ 8.48953212, 5.57319475, 8.21241294, 16.40718668, 30.15751598, 49.46340083, 74.32484124, 104.74183721, 140.71438873, 182.2424958 ],
[ 10.96088904, 3.72881146, 2.05228945, 5.93132298, 15.36591208, 30.35605673, 50.90175693, 77.00301269, 108.65982401, 145.87219088]])
prob = om.Problem()
mm = prob.model.add_subsystem('mm', om.MetaModelStructuredComp(method='slinear'),
promotes=['x0', 'x1'])
mm.add_input('x0', 0.0, train_x0)
mm.add_input('x1', 0.0, train_x1)
mm.add_output('f', 0.0, t_data)
prob.setup()
prob.final_setup()
Unstructured MetaModel Example Script#
To view this example metamodel, copy the following script into a file named unstructured_meta_model_example.py and then run openmdao view_mm unstructured_meta_model_example.py in the command line.
from math import pi
# Model
interp = om.MetaModelUnStructuredComp()
# Training Data
x_train1 = np.random.uniform(0, pi, 100)
x_train2 = np.random.uniform(0, pi, 100)
x_train3 = np.random.uniform(0, pi, 100)
y = np.sin(x_train1 * x_train2 * x_train3)
# Inputs
interp.add_input('input_1', 0., training_data=x_train1)
interp.add_input('input_2', 0., training_data=x_train2)
interp.add_input('input_3', 0., training_data=x_train3)
# Outputs
interp.add_output('output_1', 0., training_data=y)
# Surrogate Model
interp.options['default_surrogate'] = om.KrigingSurrogate()
prob = om.Problem()
prob.model.add_subsystem('interp', interp)
prob.setup()
prob.final_setup()
Note
OpenMDAO supports structured and unstructured metamodels. Please refer to the documentation for a more in depth overview of what Unstructured and Structured metamodels are.
Multiple Meta Models in Script#
If your model has multiple metamodels, you can specify which of them you want to visualize. For example, in this code there are two metamodels.
class CosMetaModel(om.MetaModelUnStructuredComp):
def setup(self):
# Training Data
x_train = np.linspace(0, 10, 20)
y_train = np.linspace(0, 20, 20)
# Inputs
self.add_input('x', 0., training_data=x_train)
self.add_input('y', 0., training_data=y_train)
# Outputs
self.add_output('cos_x', 0., training_data=np.cos(x_train + y_train))
# Surrogate Model
self.options['default_surrogate'] = om.ResponseSurface()
class SinMetaModel(om.MetaModelUnStructuredComp):
def setup(self):
# Training Data
x_train = np.linspace(0, 10, 20)
y_train = np.linspace(0, 20, 20)
# Inputs
self.add_input('x', 0., training_data=x_train)
self.add_input('y', 0., training_data=y_train)
# Outputs
self.add_output('sin_x', 0., training_data=np.sin(x_train + y_train))
# Surrogate Model
self.options['default_surrogate'] = om.ResponseSurface()
# define model with two metamodel components
model = om.Group()
cos_mm = model.add_subsystem('cos_mm', CosMetaModel())
sin_mm = model.add_subsystem('sin_mm', SinMetaModel())
# setup a problem using our dual metamodel model
prob = om.Problem(model)
prob.setup()
prob.final_setup()
To visualize only the first one, you would use the command:
openmdao view_mm -m cos_mm multiple_metamodels.py
Command Line Interface#
The command, openmdao view_mm requires a file path, the name of the meta model which you want to visualize if there is more than one, and optionally a port number:
openmdao view_mm -h
usage: openmdao view_mm [-h] [-m PATHNAME] [-r RESOLUTION] [-p PORT_NUMBER]
[--no_browser]
file
positional arguments:
file Python file containing the model.
options:
-h, --help show this help message and exit
-m, --metamodel_pathname PATHNAME
pathname of the metamodel component.
-r, --resolution RESOLUTION
Number of points to create contour grid
-p, --port_number PORT_NUMBER
Port number to open viewer
--no_browser Bokeh server will start server without browser
Note
When using Bash on Windows you are required to set –no_browser option to start the server and then open an internet browser and copy/paste the path to viewer. Bash on Windows does not allow the terminal to access your browser to open the viewer.
