MetaModelSemiStructuredComp#
Semi-structued training data occurs frequently in engineering applications like performance maps for turbine engine performance, propeller performance, aerodyanmic drag, etc. For example, a simple semi-structured grid could be a mostly structured grid, but with a few data points missing on one area of the grid. Another more general kind of semi-structured grid is a set of data where the first dimension is regular, but the other dimensions are irregular. An example of this is the figure below, where the ‘x’ dimension is regular, and the data points fall on distinct values, but the ‘y’ dimension is not regular.
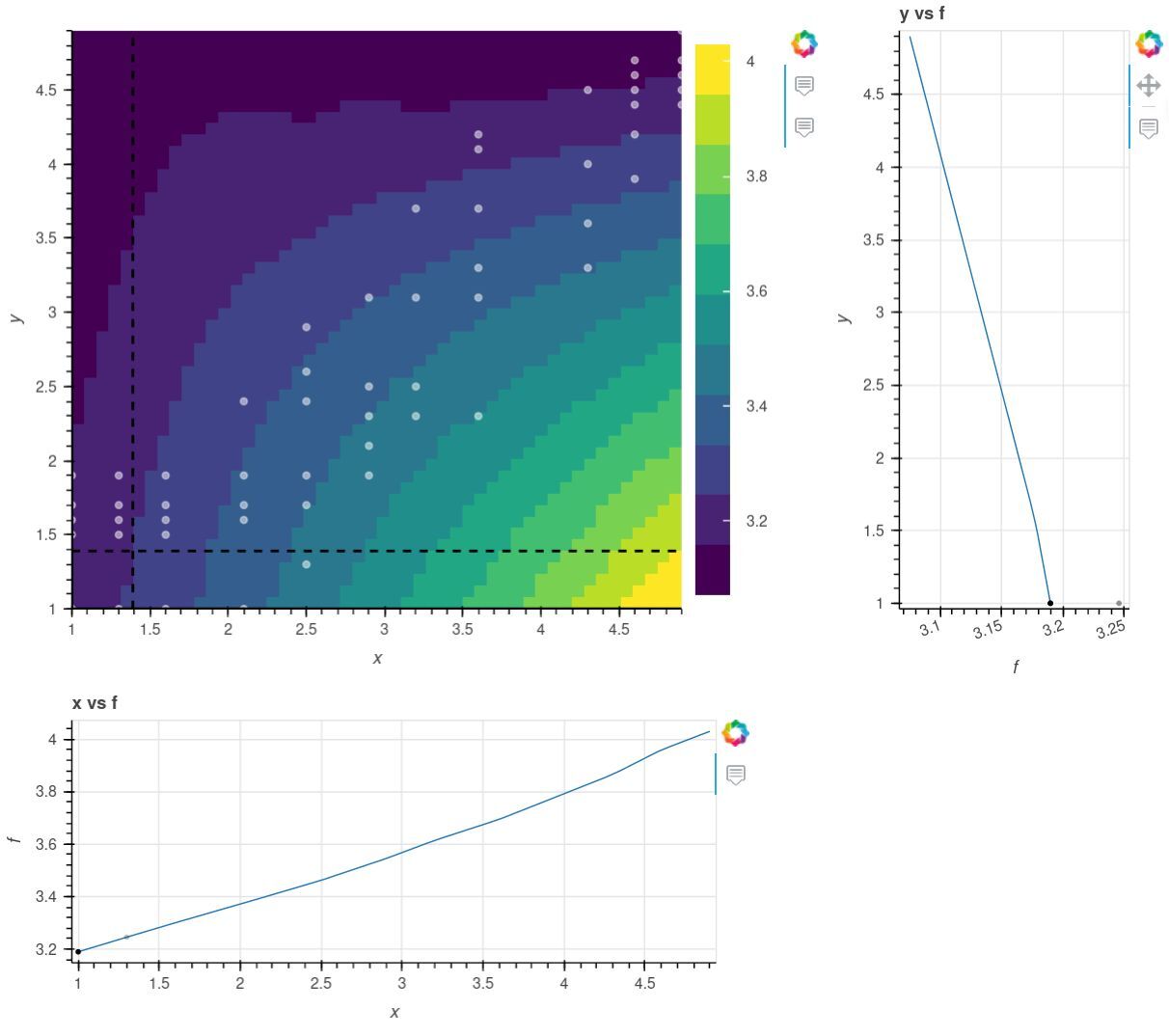
The MetaModelStructured cannot be used if any points are missing, or if any dimension is not regular. You could use MetaModelUnStructured for this data, but you would lose the performance and flexibility you can gain from exploiting the semi structured nature of the data.
MetaModelSemiStructuredComp handles semi-structured data by treating every “column” of data beyond the first dimension independently. In the figure above, it doesn’t matter that two adjacent columns have different points, as the interpolation in ‘y’ happens independently in each column before those values are subsequently interpolated in ‘x’.
The following interpolation methods are supported in MetaModelSemiStructuredComp:
Method |
Order |
Description |
|---|---|---|
slinear |
1 |
Basic linear interpolation |
lagrange2 |
2 |
Second order Lagrange polynomial |
lagrange3 |
3 |
Third order Lagrange polynomial |
akima |
3 |
Interpolation using Akima splines |
Note that MetaModelSemiStructuredComp only accepts scalar inputs and outputs. If you have a multivariable function, each input variable needs its own named OpenMDAO input.
For multi-dimensional data, fits are computed on a separable per-axis basis. A single interpolation method is used for all dimensions, so the minimum table dimension must be high enough to use the chosen interpolate. Further, the minimum dimension will potentially be driven by the sparsest area of your data, so care should be taken to choose the smallest regular dimension as the first dimension.
Extrapolation is supported, and is enabled by default. Given the more sparse nature of semi-structured data, we expect that extrapolation will occur more often. When evaluating a point, if any of the requested values falls beyond the first or last point in a row, you are extrapolating. There is always a risk when extrapolating, so it is important to be careful. Extrapolation can be disabled via the extrapolate option (see below).
MetaModelSemiStructuredComp Options#
| Option | Default | Acceptable Values | Acceptable Types | Description |
|---|---|---|---|---|
| always_opt | False | [True, False] | ['bool'] | If True, force nonlinear operations on this component to be included in the optimization loop even if this component is not relevant to the design variables and responses. |
| default_shape | (1,) | N/A | ['tuple'] | Default shape for variables that do not set val to a non-scalar value or set shape, shape_by_conn, copy_shape, or compute_shape. Default is (1,). |
| derivs_method | N/A | ['jax', 'cs', 'fd', None] | N/A | The method to use for computing derivatives |
| distributed | False | [True, False] | ['bool'] | If True, set all variables in this component as distributed across multiple processes |
| extrapolate | True | [True, False] | ['bool'] | Sets whether extrapolation should be performed when an input is out of bounds. |
| method | slinear | ['slinear', 'lagrange2', 'lagrange3', 'akima'] | N/A | Spline interpolation method to use for all outputs. |
| run_root_only | False | [True, False] | ['bool'] | If True, call compute, compute_partials, linearize, apply_linear, apply_nonlinear, solve_linear, solve_nonlinear, and compute_jacvec_product only on rank 0 and broadcast the results to the other ranks. |
| training_data_gradients | False | [True, False] | ['bool'] | When True, compute gradients with respect to training data values. |
| use_jit | True | [True, False] | ['bool'] | If True, attempt to use jit on compute_primal, assuming jax or some other AD package capable of jitting is active. |
| vec_size | 1 | N/A | ['int'] | Number of points to evaluate at once. |
MetaModelSemiStructuredComp Constructor#
The call signature for the MetaModelSemiStructuredComp constructor is:
- MetaModelSemiStructuredComp.__init__(**kwargs)[source]
Initialize all attributes.
MetaModelSemiStructuredComp Examples#
A simple quick-start example is fitting a 2-dimensional data set that has been evaluated on a small selection of semi structured points:
import numpy as np
data_x = np.array([
1.0,
1.0,
1.0,
1.0,
1.0,
1.3,
1.3,
1.3,
1.3,
1.3,
1.6,
1.6,
1.6,
1.6,
1.6,
2.1,
2.1,
2.1,
2.1,
2.1,
2.5,
2.5,
2.5,
2.5,
2.5,
2.5,
2.9,
2.9,
2.9,
2.9,
2.9,
3.2,
3.2,
3.2,
3.2,
3.6,
3.6,
3.6,
3.6,
3.6,
3.6,
4.3,
4.3,
4.3,
4.3,
4.6,
4.6,
4.6,
4.6,
4.6,
4.6,
4.9,
4.9,
4.9,
4.9,
4.9,
])
data_y = np.array([
1.0,
1.5,
1.6,
1.7,
1.9,
1.0,
1.5,
1.6,
1.7,
1.9,
1.0,
1.5,
1.6,
1.7,
1.9,
1.0,
1.6,
1.7,
1.9,
2.4,
1.3,
1.7,
1.9,
2.4,
2.6,
2.9,
1.9,
2.1,
2.3,
2.5,
3.1,
2.3,
2.5,
3.1,
3.7,
2.3,
3.1,
3.3,
3.7,
4.1,
4.2,
3.3,
3.6,
4.0,
4.5,
3.9,
4.2,
4.4,
4.5,
4.6,
4.7,
4.4,
4.5,
4.6,
4.7,
4.9,
])
data_values = 3.0 + np.sin(data_x*0.2) * np.cos(data_y*0.3)
Notice that the table input data is expressed as flat ndarrays (or lists). They should be sorted in ascending coordinate order starting with the lowest n-tuple. The table output values are also in a flat array that corresponds to the input arrays.
import openmdao.api as om
prob = om.Problem()
model = prob.model
interp = om.MetaModelSemiStructuredComp(method='lagrange2')
# The order that the inputs are added matters. The first table dimension is 'x'.
interp.add_input('x', data_x)
interp.add_input('y', data_y)
interp.add_output('f', training_data=data_values)
# Including a second output to show that multiple outputs are supported.
# All outputs use the same input grid but use different values at each point.
interp.add_output('g', training_data=2.0 * data_values)
model.add_subsystem('interp', interp)
prob.setup()
prob.set_val('interp.x', np.array([3.1]))
prob.set_val('interp.y', np.array([2.75]))
prob.run_model()
print(prob.get_val('interp.f'), prob.get_val('interp.g'))
[3.39415716] [6.78831431]
You can also predict multiple independent output points by setting the vec_size argument to be equal to the number of points you want to predict. Here, we set it to 3 and predict 3 points with MetaModelSemiStructuredComp:
prob = om.Problem()
model = prob.model
interp = om.MetaModelSemiStructuredComp(method='lagrange2', vec_size=3)
# The order that the inputs are added matters. The first table dimension is 'x'.
interp.add_input('x', data_x)
interp.add_input('y', data_y)
interp.add_output('f', training_data=data_values)
model.add_subsystem('interp', interp)
prob.setup()
prob.set_val('interp.x', np.array([3.1, 4.7, 5.1]))
prob.set_val('interp.y', np.array([2.75, 3.9, 4.2]))
prob.run_model()
print(prob.get_val('interp.f'))
[3.39415716 3.31511313 3.2608656 ]
Finally, it is possible to compute gradients with respect to the given
output training data. These gradients are not computed by default, but
can be enabled by setting the option training_data_gradients to True.
When this is done, for each output that is added to the component, a
corresponding input is added to the component with the same name but with an
_train suffix. This allows you to connect in the training data as an input
array, if desired.
The following simple example shows the preceding model with training inputs enabled.
prob = om.Problem()
model = prob.model
interp = om.MetaModelSemiStructuredComp(method='lagrange2', training_data_gradients=True)
interp.add_input('x', data_x)
interp.add_input('y', data_y)
# Initialize all trable values to zero.
interp.add_output('f', training_data=np.zeros(len(data_x)))
model.add_subsystem('interp', interp)
prob.setup(force_alloc_complex=True)
prob.set_val('interp.x', np.array([3.1]))
prob.set_val('interp.y', np.array([2.75]))
# The table values come from somewhere in OpenMDAO, in this case, from the indepvarcomp output.
# They could also come from an upstream component if you connect the input to another source.
prob.set_val('interp.f_train', data_values)
prob.run_model()
print(prob.get_val('interp.f'))
[3.39415716]
Standalone Interface for Table Interpolation#
The underlying interpolation algorithms can be used standalone (i.e., outside of the
MetaModelSemiStructuredComp) through the InterpNDSemi class. This can be useful for inclusion in another
component. The following component shows how to perform interpolation on the a much simpler table than before. We only have 2 points at x=1.0, so we can only choose ‘slinear’.
from openmdao.components.interp_util.interp_semi import InterpNDSemi
# Simple grid with one point missing.
x = [1.0, 1.0, 2.0, 2.0, 2.0]
y = [1.0, 2.0, 1.0, 2.0, 3.0]
values = [1.0, 2.5, 1.5, 4.0, 4.5]
grid = np.array([x, y]).T
print(grid)
# We only have 2 points at x=1.0, so 'slinear' is the only one we can use.
interp = InterpNDSemi(grid, values, method='slinear')
x = np.array([1.5, 1.5])
f, df_dx = interp.interpolate(x, compute_derivative=True)
print('value', f)
print('derivative', df_dx)
[[1. 1.]
[1. 2.]
[2. 1.]
[2. 2.]
[2. 3.]]
value [2.25]
derivative [[1. 2.]]
