The Van der Pol Oscillator#
In dynamics, the Van Der Pol oscillator [Wikipediacontributors20] is a non-conservative oscillator with non-linear damping. It evolves in time according to the second-order differential equation:
where \(x\) is the position coordinate (a function of the time \(t\)), and \(u\) is a scalar parameter indicating the nonlinearity and the strength of the damping.
To make this an optimal control problem, we want to find the smallest control that will dampen the oscillation (drive the state variables to zero). We can express this as an objective function \(J\) to minimize:
In other words, we want to find the optimal (smallest) trajectory of the control \(u\) such that the oscillation and the oscillation’s rate of change are driven to zero.
State Variables#
There are three state variables are used to define the configuration of the system at any given instant in time.
\(x_1\): The primary output of the oscillator.
\(x_0\): The rate of change of the primary output.
\(J\): The objective function to be minimized.
The objective function is included as a state variable so that Dymos will do the integration.
The \(x_1\) and \(x_0\) state variables are also inputs to the system, along with the control \(u\).
System Dynamics#
The evolution of the state variables is given by the following ordinary differential equations (ODE):
Control Variables#
This system has a single control variable:
\(u\): The control input.
The control variable has a constraint: \(-0.75 \leq u \leq 1.0\)
The initial and final conditions#
The initial conditions are:
The final conditions are:
Defining the ODE as an OpenMDAO System#
In Dymos, the ODE is an OpenMDAO System (a Component, or a Group of components). The following ExplicitComponent computes the state rates for the Van der Pol problem.
More detail on the workings of an ExplicitComponent can be found in the OpenMDAO documentation. In summary:
initialize: Called at setup, and used to define options for the component. ALL Dymos ODE components should have the property
num_nodes, which defines the number of points at which the outputs are simultaneously computed.setup: Used to add inputs and outputs to the component, and declare which outputs (and indices of outputs) are dependent on each of the inputs.
compute: Used to compute the outputs, given the inputs.
compute_partials: Used to compute the derivatives of the outputs with respect to each of the inputs analytically. This method may be omitted if finite difference or complex-step approximations are used, though analytic is recommended.
Note
Things to note about the Van der Pol ODE system
Only the vanderpol_ode class below is important for defining the basic problem. The other classes are used to demonstrate Message Passing Interface (MPI) parallel calculation of the system. They can be ignored.
\(x_1\), \(x_0\), and \(u\) are inputs.
\(\dot{x_1}\), \(\dot{x_0}\), and \(\dot{J}\) are outputs.
declare_partials is called for every output with respect to every input.
For efficiency, partial derrivatives that are constant have values specified in the setup method rather than the compute_partials method. So although 7 partials are declared, only 5 are computed in compute_partials.
This ODE includes some additional complexity to allow it to run in parallel. See the Van der Pol example in the dymos examples directory for an example of how to run the problem under MPI.
import numpy as np
import openmdao.api as om
from openmdao.utils.array_utils import evenly_distrib_idxs
class VanderpolODE(om.ExplicitComponent):
"""intentionally slow version of vanderpol_ode for effects of demonstrating distributed component calculations
MPI can run this component in multiple processes, distributing the calculation of derivatives.
This code has a delay in it to simulate a longer computation. It should run faster with more processes.
"""
def __init__(self, *args, **kwargs):
self.progress_prints = False
super().__init__(*args, **kwargs)
def initialize(self):
self.options.declare('num_nodes', types=int)
self.options.declare('distrib', types=bool, default=False)
def setup(self):
nn = self.options['num_nodes']
comm = self.comm
rank = comm.rank
sizes, offsets = evenly_distrib_idxs(comm.size, nn) # (#cpus, #inputs) -> (size array, offset array)
self.start_idx = offsets[rank]
self.io_size = sizes[rank] # number of inputs and outputs managed by this distributed process
self.end_idx = self.start_idx + self.io_size
# inputs: 2 states and a control
self.add_input('x0', val=np.ones(nn), desc='derivative of Output', units='V/s')
self.add_input('x1', val=np.ones(nn), desc='Output', units='V')
self.add_input('u', val=np.ones(nn), desc='control', units=None)
# outputs: derivative of states
# the objective function will be treated as a state for computation, so its derivative is an output
self.add_output('x0dot', val=np.ones(self.io_size), desc='second derivative of Output',
units='V/s**2', distributed=self.options['distrib'])
self.add_output('x1dot', val=np.ones(self.io_size), desc='derivative of Output',
units='V/s', distributed=self.options['distrib'])
self.add_output('Jdot', val=np.ones(self.io_size), desc='derivative of objective',
units='1.0/s', distributed=self.options['distrib'])
# self.declare_coloring(method='cs')
# # partials
r = np.arange(self.io_size, dtype=int)
c = r + self.start_idx
self.declare_partials(of='x0dot', wrt='x0', rows=r, cols=c)
self.declare_partials(of='x0dot', wrt='x1', rows=r, cols=c)
self.declare_partials(of='x0dot', wrt='u', rows=r, cols=c, val=1.0)
self.declare_partials(of='x1dot', wrt='x0', rows=r, cols=c, val=1.0)
self.declare_partials(of='Jdot', wrt='x0', rows=r, cols=c)
self.declare_partials(of='Jdot', wrt='x1', rows=r, cols=c)
self.declare_partials(of='Jdot', wrt='u', rows=r, cols=c)
def compute(self, inputs, outputs):
# The inputs contain the entire vector, be each rank will only operate on a portion of it.
x0 = inputs['x0'][self.start_idx:self.end_idx]
x1 = inputs['x1'][self.start_idx:self.end_idx]
u = inputs['u'][self.start_idx:self.end_idx]
outputs['x0dot'] = (1.0 - x1**2) * x0 - x1 + u
outputs['x1dot'] = x0
outputs['Jdot'] = x0**2 + x1**2 + u**2
def compute_partials(self, inputs, jacobian):
x0 = inputs['x0'][self.start_idx:self.end_idx]
x1 = inputs['x1'][self.start_idx:self.end_idx]
u = inputs['u'][self.start_idx:self.end_idx]
jacobian['x0dot', 'x0'] = 1.0 - x1 * x1
jacobian['x0dot', 'x1'] = -2.0 * x1 * x0 - 1.0
jacobian['Jdot', 'x0'] = 2.0 * x0
jacobian['Jdot', 'x1'] = 2.0 * x1
jacobian['Jdot', 'u'] = 2.0 * u
Defining the Dymos Problem#
Once the ODEs are defined, they are used to create a Dymos Problem object that allows solution.
Note
Things to note about the Van der Pol Dymos Problem definition
The vanderpol function creates and returns a Dymos Problem instance that can be used for simulation or optimization.
The vanderpol function has optional arguments for specifying options for the type of transcription, number of segments, optimizer, etc. These can be ignored when first trying to understand the code.
The Problem object has a Trajectory object, and the trajectory has a single Phase. Most of the problem setup is performed by calling methods on the phase (set_time_options, add_state, add_boundary_constraint, add_objective).
The add_state and add_control calls include the target parameter for \(x_0\), \(x_1\), and \(u\). This is required so that the inputs are correctly calculated.
Initial (linear) guesses are supplied for the states and control.
import openmdao.api as om
import dymos as dm
def vanderpol(transcription='gauss-lobatto', num_segments=15, transcription_order=3,
compressed=True, optimizer='SLSQP', use_pyoptsparse=False):
"""Dymos problem definition for optimal control of a Van der Pol oscillator"""
# define the OpenMDAO problem
p = om.Problem(model=om.Group())
if not use_pyoptsparse:
p.driver = om.ScipyOptimizeDriver()
else:
p.driver = om.pyOptSparseDriver(print_results=False)
p.driver.options['optimizer'] = optimizer
if use_pyoptsparse:
if optimizer == 'SNOPT':
p.driver.opt_settings['iSumm'] = 6 # show detailed SNOPT output
elif optimizer == 'IPOPT':
p.driver.opt_settings['print_level'] = 4
p.driver.declare_coloring()
# define a Trajectory object and add to model
traj = dm.Trajectory()
p.model.add_subsystem('traj', subsys=traj)
# define a Transcription
if transcription == 'gauss-lobatto':
t = dm.GaussLobatto(num_segments=num_segments,
order=transcription_order,
compressed=compressed)
elif transcription == 'radau-ps':
t = dm.Radau(num_segments=num_segments,
order=transcription_order,
compressed=compressed)
# define a Phase as specified above and add to Phase
phase = dm.Phase(ode_class=VanderpolODE, transcription=t)
traj.add_phase(name='phase0', phase=phase)
t_final = 10
phase.set_time_options(fix_initial=True, fix_duration=True, duration_val=t_final, units='s')
# set the State time options
phase.add_state('x0', fix_initial=True, fix_final=True,
rate_source='x0dot',
units='V/s', ref=0.1, defect_ref=0.1) # target required because x0 is an input
phase.add_state('x1', fix_initial=True, fix_final=True,
rate_source='x1dot',
units='V', ref=0.1, defect_ref=0.1)
phase.add_state('J', fix_initial=True, fix_final=False,
rate_source='Jdot',
units=None)
# define the control
phase.add_control(name='u', units=None, lower=-0.75, upper=1.0, continuity=True,
rate_continuity=True)
# define objective to minimize
phase.add_objective('J', loc='final')
# setup the problem
p.setup(check=True)
phase.set_time_val(0.0, t_final)
# add a linearly interpolated initial guess for the state and control curves
phase.set_state_val('x0', [1, 0])
phase.set_state_val('x0', [1, 0])
phase.set_state_val('x0', [0, 1])
phase.set_control_val('u', -0.75)
return p
Simulating the Problem (without control)#
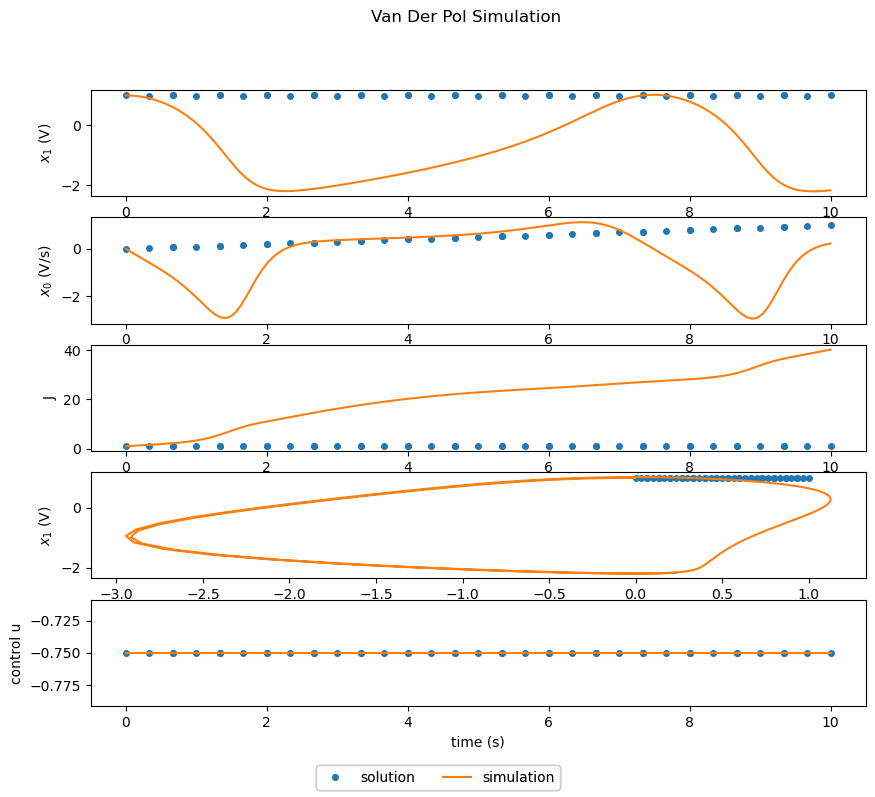
The following script creates an instance of the Dymos vanderpol problem and simulates it.
Since the problem was only simulated and not solved, the solution lines in the plots show only the initial guesses for \(x_0\), \(x_1\), and \(u\). The simulation lines shown in the plots are the system response with the control variable \(u\) held constant.
# Create the Dymos problem instance
p = vanderpol(transcription='gauss-lobatto', use_pyoptsparse=True, optimizer='IPOPT', num_segments=15)
# Run the problem (simulate only)
dm.run_problem(p, run_driver=False, simulate=True)
INFO: checking out_of_order...
INFO: out_of_order check complete (0.000329 sec).
INFO: checking system...
INFO: system check complete (0.000026 sec).
INFO: checking solvers...
INFO: solvers check complete (0.000198 sec).
INFO: checking dup_inputs...
INFO: dup_inputs check complete (0.000118 sec).
INFO: checking missing_recorders...
INFO: missing_recorders check complete (0.000002 sec).
INFO: checking unserializable_options...
INFO: unserializable_options check complete (0.001225 sec).
INFO: checking comp_has_no_outputs...
INFO: comp_has_no_outputs check complete (0.000033 sec).
INFO: checking auto_ivc_warnings...
INFO: auto_ivc_warnings check complete (0.000002 sec).
Simulating trajectory traj
Done simulating trajectory traj
The first two plots shows the variables \(x_0\) and \(x_1\) vs time. The third plots shows \(x_0\) vs. \(x_1\) (which will be mostly circular in the case of undamped oscillation). The final plot is the (fixed) control variable \(u\) vs time.
(<Figure size 1000x800 with 5 Axes>,
array([<Axes: xlabel='time (s)', ylabel='$x_1$ (V)'>,
<Axes: xlabel='time (s)', ylabel='$x_0$ (V/s)'>,
<Axes: xlabel='time (s)', ylabel='J'>,
<Axes: xlabel='$x_0$ (V/s)', ylabel='$x_1$ (V)'>,
<Axes: xlabel='time (s)', ylabel='control u'>], dtype=object))
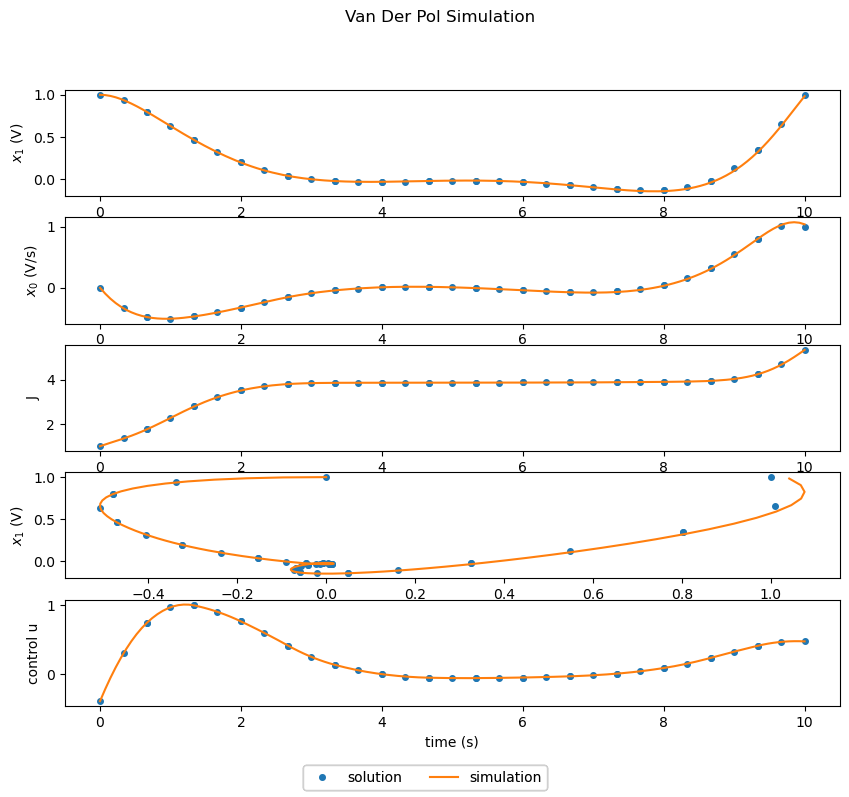
Solving the Optimal Control Problem#
The next example shows optimization followed by simulation.
With a successful optimization, the resulting plots show a good match between the simulated (with varying control) and optimized results. The state variables \(x_0\) and \(x_1\) as well as the control variable \(u\) are all driven to zero.
However one can notice that the distribution of segments/nodes in the phase results in a bit of error. If we take the simulation results as the truth, there is a bit of divergence in the state values of the solution and simulation towards the end.
import dymos as dm
# Create the Dymos problem instance
p = vanderpol(transcription='gauss-lobatto', num_segments=15,
transcription_order=3, compressed=False, use_pyoptsparse=True, optimizer='IPOPT')
# Find optimal control solution to stop oscillation
dm.run_problem(p, simulate=True)
INFO: checking out_of_order...
INFO: out_of_order check complete (0.000437 sec).
INFO: checking system...
INFO: system check complete (0.000019 sec).
INFO: checking solvers...
INFO: solvers check complete (0.000160 sec).
INFO: checking dup_inputs...
INFO: dup_inputs check complete (0.000091 sec).
INFO: checking missing_recorders...
INFO: missing_recorders check complete (0.000002 sec).
INFO: checking unserializable_options...
INFO: unserializable_options check complete (0.001133 sec).
INFO: checking comp_has_no_outputs...
INFO: comp_has_no_outputs check complete (0.000030 sec).
INFO: checking auto_ivc_warnings...
INFO: auto_ivc_warnings check complete (0.000003 sec).
Jacobian shape: (60, 130) (5.15% nonzero)
FWD solves: 0 REV solves: 5
Total colors vs. total size: 5 vs 60 (91.67% improvement)
Sparsity computed using tolerance: 1e-25.
Dense total jacobian for Problem 'problem4' was computed 3 times.
Time to compute sparsity: 0.0778 sec
Time to compute coloring: 0.0540 sec
Memory to compute coloring: 0.3086 MB
Coloring created on: 2026-02-26 12:24:18
List of user-set options:
Name Value used
file_print_level = 5 yes
hessian_approximation = limited-memory yes
linear_solver = mumps yes
nlp_scaling_method = user-scaling yes
output_file = /home/runner/work/dymos/dymos/docs/dymos_book/examples/vanderpol/problem4_out/IPOPT.out yes
print_level = 4 yes
print_user_options = yes yes
sb = yes yes
Total number of variables............................: 130
variables with only lower bounds: 0
variables with lower and upper bounds: 45
variables with only upper bounds: 0
Total number of equality constraints.................: 115
Total number of inequality constraints...............: 0
inequality constraints with only lower bounds: 0
inequality constraints with lower and upper bounds: 0
inequality constraints with only upper bounds: 0
Number of Iterations....: 31
(scaled) (unscaled)
Objective...............: 5.3660057662482830e+00 5.3660057662482830e+00
Dual infeasibility......: 5.6619848758030917e-09 5.6619848758030917e-09
Constraint violation....: 7.7715611723760958e-15 7.7715611723760958e-15
Variable bound violation: 9.8138401849467982e-09 9.8138401849467982e-09
Complementarity.........: 1.0000001534440038e-11 1.0000001534440038e-11
Overall NLP error.......: 5.6619848758030917e-09 5.6619848758030917e-09
Number of objective function evaluations = 33
Number of objective gradient evaluations = 32
Number of equality constraint evaluations = 33
Number of inequality constraint evaluations = 0
Number of equality constraint Jacobian evaluations = 32
Number of inequality constraint Jacobian evaluations = 0
Number of Lagrangian Hessian evaluations = 0
Total seconds in IPOPT = 0.734
EXIT: Optimal Solution Found.
/home/runner/work/dymos/dymos/.openmdao-pixi/.pixi/envs/dev/lib/python3.13/site-packages/openmdao/visualization/opt_report/opt_report.py:611: UserWarning: Attempting to set identical low and high ylims makes transformation singular; automatically expanding.
ax.set_ylim([ymin_plot, ymax_plot])
Simulating trajectory traj
Done simulating trajectory traj
Problem: problem4
Driver: pyOptSparseDriver
success : True
iterations : 35
runtime : 9.0415E-01 s
model_evals : 36
model_time : 2.6674E-02 s
deriv_evals : 33
deriv_time : 1.6175E-01 s
exit_status : SUCCESS
(<Figure size 1000x800 with 5 Axes>,
array([<Axes: xlabel='time (s)', ylabel='$x_1$ (V)'>,
<Axes: xlabel='time (s)', ylabel='$x_0$ (V/s)'>,
<Axes: xlabel='time (s)', ylabel='J'>,
<Axes: xlabel='$x_0$ (V/s)', ylabel='$x_1$ (V)'>,
<Axes: xlabel='time (s)', ylabel='control u'>], dtype=object))
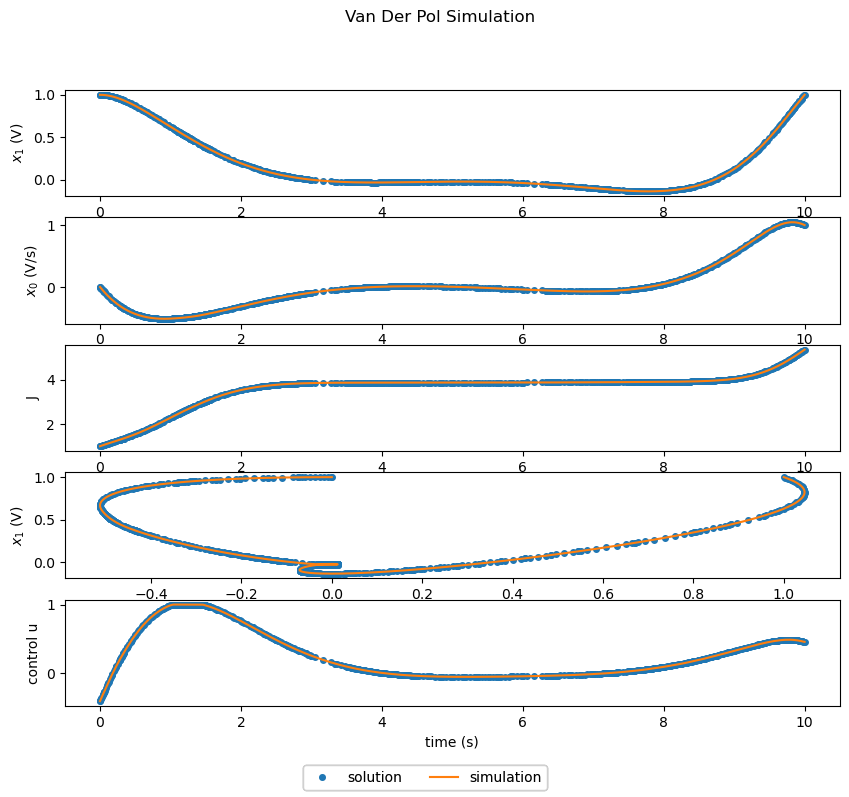
Solving the Optimal Control Problem with Grid Refinement#
Repeating the optimization with grid refinement enabled requires changing only two lines in the code. For the sake of grid refinement demonstration, the initial number of segments is also reduced by a factor of 5.
Optimization with grid refinement gets results similar to the example without grid refinement, but runs faster and does not require supplying a good guess for the number segments.
# Create the Dymos problem instance
p = vanderpol(transcription='gauss-lobatto', num_segments=15,
transcription_order=3, compressed=True, optimizer='SLSQP')
# Enable grid refinement and find optimal control solution to stop oscillation
p.model.traj.phases.phase0.set_refine_options(refine=True, tol=1.0E-6)
dm.run_problem(p, simulate=True, refine_iteration_limit=5, refine_method='hp')
INFO: checking out_of_order...
INFO: checking out_of_order...
INFO: out_of_order check complete (0.000297 sec).
INFO: out_of_order check complete (0.000297 sec).
INFO: checking system...
INFO: checking system...
INFO: system check complete (0.000014 sec).
INFO: system check complete (0.000014 sec).
INFO: checking solvers...
INFO: checking solvers...
INFO: solvers check complete (0.000169 sec).
INFO: solvers check complete (0.000169 sec).
INFO: checking dup_inputs...
INFO: checking dup_inputs...
INFO: dup_inputs check complete (0.000051 sec).
INFO: dup_inputs check complete (0.000051 sec).
INFO: checking missing_recorders...
INFO: checking missing_recorders...
INFO: missing_recorders check complete (0.000001 sec).
INFO: missing_recorders check complete (0.000001 sec).
INFO: checking unserializable_options...
INFO: checking unserializable_options...
INFO: unserializable_options check complete (0.001922 sec).
INFO: unserializable_options check complete (0.001922 sec).
INFO: checking comp_has_no_outputs...
INFO: checking comp_has_no_outputs...
INFO: comp_has_no_outputs check complete (0.000037 sec).
INFO: comp_has_no_outputs check complete (0.000037 sec).
INFO: checking auto_ivc_warnings...
INFO: checking auto_ivc_warnings...
INFO: auto_ivc_warnings check complete (0.000001 sec).
INFO: auto_ivc_warnings check complete (0.000001 sec).
Jacobian shape: (60, 74) (8.74% nonzero)
FWD solves: 9 REV solves: 0
Total colors vs. total size: 9 vs 74 (87.84% improvement)
Sparsity computed using tolerance: 1e-25.
Dense total jacobian for Problem 'problem7' was computed 3 times.
Time to compute sparsity: 0.0762 sec
Time to compute coloring: 0.0528 sec
Memory to compute coloring: 0.0273 MB
Coloring created on: 2026-02-26 12:24:21
Optimization terminated successfully (Exit mode 0)
Current function value: 5.3660108149594405
Iterations: 43
Function evaluations: 45
Gradient evaluations: 43
Optimization Complete
-----------------------------------
==================================================
Grid Refinement - Iteration 1
--------------------------------------------------
Phase: traj.phases.phase0
Refinement Options:
Allow Refinement = True
Tolerance = 1e-06
Min Order = 3
Max Order = 14
Original Grid:
Number of Segments = 15
Segment Ends = [-1.0, -0.8667, -0.7333, -0.6, -0.4667, -0.3333, -0.2, -0.0667, 0.0667, 0.2, 0.3333, 0.4667, 0.6, 0.7333, 0.8667, 1.0]
Segment Order = [3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3, 3]
Error = [ 0.02474, 0.01233, 0.00217, 0.004157, 0.002431, 0.001684, 0.001182, 0.0006564, 0.0008722, 0.001531, 0.002867, 0.004009, 0.006365, 0.01045, 0.03593]
Phase: traj.phases.phase0
New Grid:
Number of Segments = 15
Segment Ends = [-1.0, -0.8667, -0.7333, -0.6, -0.4667, -0.3333, -0.2, -0.0667, 0.0667, 0.2, 0.3333, 0.4667, 0.6, 0.7333, 0.8667, 1.0]
Segment Order = [5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5]
Refined: True
/home/runner/work/dymos/dymos/.openmdao-pixi/.pixi/envs/dev/lib/python3.13/site-packages/openmdao/core/system.py:4656: OpenMDAOWarning:Calling `list_inputs` before `final_setup` will only display the default values of variables and will not show the result of any `set_val` calls.
/home/runner/work/dymos/dymos/.openmdao-pixi/.pixi/envs/dev/lib/python3.13/site-packages/dymos/phase/phase.py:3051: OpenMDAOWarning:phase0.states:x0 specifies 'fix_final=True'. If the given restart file has a different final value this will overwrite the user-specified value
/home/runner/work/dymos/dymos/.openmdao-pixi/.pixi/envs/dev/lib/python3.13/site-packages/dymos/phase/phase.py:3051: OpenMDAOWarning:phase0.states:x1 specifies 'fix_final=True'. If the given restart file has a different final value this will overwrite the user-specified value
Jacobian shape: (105, 149) (6.78% nonzero)
FWD solves: 14 REV solves: 0
Total colors vs. total size: 14 vs 149 (90.60% improvement)
Sparsity computed using tolerance: 1e-25.
Dense total jacobian for Problem 'problem7' was computed 3 times.
Time to compute sparsity: 0.1475 sec
Time to compute coloring: 0.0964 sec
Memory to compute coloring: 0.0000 MB
Coloring created on: 2026-02-26 12:24:22
Optimization terminated successfully (Exit mode 0)
Current function value: 5.358596871051768
Iterations: 20
Function evaluations: 21
Gradient evaluations: 20
Optimization Complete
-----------------------------------
==================================================
Grid Refinement - Iteration 2
--------------------------------------------------
Phase: traj.phases.phase0
Refinement Options:
Allow Refinement = True
Tolerance = 1e-06
Min Order = 3
Max Order = 14
Original Grid:
Number of Segments = 15
Segment Ends = [-1.0, -0.8667, -0.7333, -0.6, -0.4667, -0.3333, -0.2, -0.0667, 0.0667, 0.2, 0.3333, 0.4667, 0.6, 0.7333, 0.8667, 1.0]
Segment Order = [5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5]
Error = [0.0003551, 0.001424, 0.00034, 2.618e-05, 3.937e-05, 4.898e-05, 5.822e-06, 7.636e-06, 2.675e-06, 5.192e-06, 9.963e-06, 7.221e-06, 5.45e-05, 0.0002363, 0.00118]
Phase: traj.phases.phase0
New Grid:
Number of Segments = 33
Segment Ends = [-1.0, -0.9333, -0.8667, -0.8222, -0.7778, -0.7333, -0.6889, -0.6444, -0.6, -0.5333, -0.4667, -0.4, -0.3333, -0.2667, -0.2, -0.1333, -0.0667, 0.0, 0.0667, 0.1333, 0.2, 0.2667, 0.3333, 0.4, 0.4667, 0.5333, 0.6, 0.6667, 0.7333, 0.8, 0.8667, 0.9111, 0.9556, 1.0]
Segment Order = [5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5]
Refined: True
/home/runner/work/dymos/dymos/.openmdao-pixi/.pixi/envs/dev/lib/python3.13/site-packages/openmdao/core/system.py:4656: OpenMDAOWarning:Calling `list_inputs` before `final_setup` will only display the default values of variables and will not show the result of any `set_val` calls.
/home/runner/work/dymos/dymos/.openmdao-pixi/.pixi/envs/dev/lib/python3.13/site-packages/dymos/phase/phase.py:3051: OpenMDAOWarning:phase0.states:x0 specifies 'fix_final=True'. If the given restart file has a different final value this will overwrite the user-specified value
/home/runner/work/dymos/dymos/.openmdao-pixi/.pixi/envs/dev/lib/python3.13/site-packages/dymos/phase/phase.py:3051: OpenMDAOWarning:phase0.states:x1 specifies 'fix_final=True'. If the given restart file has a different final value this will overwrite the user-specified value
/home/runner/work/dymos/dymos/.openmdao-pixi/.pixi/envs/dev/lib/python3.13/site-packages/openmdao/core/driver.py:700: DriverWarning:The following design variable initial conditions are out of their specified bounds:
traj.phase0.controls:u
val: [-0.41617723 -0.28112219 -0.04338289 0.17394989 0.28859841 0.37721951
0.54521354 0.68986226 0.75513804 0.79103952 0.85909594 0.90302672
0.92545988 0.94789304 0.99041832 0.99361221 0.99529707 0.99698193
1. 1. 1. 0.99903555 0.99720729 0.97461374
0.96205691 0.94950008 0.92569679 0.89049665 0.87192769 0.85335872
0.81796879 0.77932114 0.75893353 0.72676097 0.66567059 0.60446556
0.57217836 0.54146544 0.48324468 0.42691874 0.39810063 0.37100767
0.32212336 0.27600164 0.25167127 0.23090073 0.19152718 0.15523944
0.13755472 0.12184059 0.09426875 0.06917172 0.0559324 0.04594257
0.02700542 0.01000242 0.00194675 -0.00497971 -0.01659773 -0.0265275
-0.0317657 -0.03524717 -0.04184679 -0.04735861 -0.04975226 -0.05146338
-0.0539965 -0.0557363 -0.05665409 -0.05685798 -0.05724449 -0.05722883
-0.05703056 -0.05661726 -0.05564332 -0.05445671 -0.05383075 -0.05298344
-0.05137724 -0.04961481 -0.04861126 -0.04749294 -0.0452096 -0.04274383
-0.04144307 -0.03981084 -0.03671672 -0.03324111 -0.0312274 -0.02892447
-0.02410408 -0.0187758 -0.01596501 -0.01224427 -0.00519109 0.00265378
0.0071662 0.01232616 0.02308365 0.03493089 0.0411806 0.04901174
0.06385679 0.0801312 0.08939171 0.0996846 0.12050857 0.14279777
0.15455589 0.16802606 0.19356071 0.2198732 0.23412122 0.24890598
0.27783212 0.30776254 0.3235516 0.33957317 0.36994438 0.3992238
0.41415359 0.42334855 0.44077892 0.45321558 0.45962276 0.46602995
0.47817568 0.47687502 0.47618889 0.47550275 0.47344362 0.45836529
0.4504111 ]
lower: [-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75]
upper: [1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.
1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.
1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.
1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.
1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.
1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.]
Set the initial value of the design variable to a valid value or set the driver option['invalid_desvar_behavior'] to 'ignore'.
Jacobian shape: (231, 329) (3.13% nonzero)
FWD solves: 14 REV solves: 0
Total colors vs. total size: 14 vs 329 (95.74% improvement)
Sparsity computed using tolerance: 1e-25.
Dense total jacobian for Problem 'problem7' was computed 3 times.
Time to compute sparsity: 0.3383 sec
Time to compute coloring: 0.2120 sec
Memory to compute coloring: 0.0234 MB
Coloring created on: 2026-02-26 12:24:23
Optimization terminated successfully (Exit mode 0)
Current function value: 5.358606209910723
Iterations: 12
Function evaluations: 13
Gradient evaluations: 12
Optimization Complete
-----------------------------------
==================================================
Grid Refinement - Iteration 3
--------------------------------------------------
Phase: traj.phases.phase0
Refinement Options:
Allow Refinement = True
Tolerance = 1e-06
Min Order = 3
Max Order = 14
Original Grid:
Number of Segments = 33
Segment Ends = [-1.0, -0.9333, -0.8667, -0.8222, -0.7778, -0.7333, -0.6889, -0.6444, -0.6, -0.5333, -0.4667, -0.4, -0.3333, -0.2667, -0.2, -0.1333, -0.0667, 0.0, 0.0667, 0.1333, 0.2, 0.2667, 0.3333, 0.4, 0.4667, 0.5333, 0.6, 0.6667, 0.7333, 0.8, 0.8667, 0.9111, 0.9556, 1.0]
Segment Order = [5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5]
Error = [6.072e-05, 5.85e-05, 1.282e-05, 6.101e-05, 8.486e-06, 7.452e-05, 1.691e-05, 4.747e-06, 7.463e-06, 1.189e-06, 5.144e-06, 8.896e-07, 3.813e-06, 6.997e-06, 1.754e-06, 5.023e-06, 2.452e-06, 1.816e-06, 1.209e-06, 8.82e-07, 6.724e-07, 2.307e-06, 1.898e-06, 5.16e-06, 3.68e-06, 9.54e-06, 4.639e-07, 1.991e-05, 1.826e-05, 4.453e-06, 8.512e-06, 1.08e-05, 4.376e-05]
Phase: traj.phases.phase0
New Grid:
Number of Segments = 63
Segment Ends = [-1.0, -0.9778, -0.9556, -0.9333, -0.9111, -0.8889, -0.8667, -0.8444, -0.8222, -0.8074, -0.7926, -0.7778, -0.7333, -0.7185, -0.7037, -0.6889, -0.6667, -0.6444, -0.6222, -0.6, -0.5667, -0.5333, -0.4667, -0.4333, -0.4, -0.3333, -0.3, -0.2667, -0.2333, -0.2, -0.1667, -0.1333, -0.1, -0.0667, -0.0333, 0.0, 0.0333, 0.0667, 0.1333, 0.2, 0.2667, 0.3, 0.3333, 0.3667, 0.4, 0.4333, 0.4667, 0.5, 0.5333, 0.5667, 0.6, 0.6667, 0.7, 0.7333, 0.7667, 0.8, 0.8667, 0.8889, 0.9111, 0.9333, 0.9556, 0.9704, 0.9852, 1.0]
Segment Order = [5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 15, 5, 5, 5, 5, 5, 5, 5, 5, 5, 7, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 9, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 9, 5, 5, 5, 5, 5, 5, 5]
Refined: True
/home/runner/work/dymos/dymos/.openmdao-pixi/.pixi/envs/dev/lib/python3.13/site-packages/openmdao/core/system.py:4656: OpenMDAOWarning:Calling `list_inputs` before `final_setup` will only display the default values of variables and will not show the result of any `set_val` calls.
/home/runner/work/dymos/dymos/.openmdao-pixi/.pixi/envs/dev/lib/python3.13/site-packages/dymos/phase/phase.py:3051: OpenMDAOWarning:phase0.states:x0 specifies 'fix_final=True'. If the given restart file has a different final value this will overwrite the user-specified value
/home/runner/work/dymos/dymos/.openmdao-pixi/.pixi/envs/dev/lib/python3.13/site-packages/dymos/phase/phase.py:3051: OpenMDAOWarning:phase0.states:x1 specifies 'fix_final=True'. If the given restart file has a different final value this will overwrite the user-specified value
/home/runner/work/dymos/dymos/.openmdao-pixi/.pixi/envs/dev/lib/python3.13/site-packages/openmdao/core/driver.py:700: DriverWarning:The following design variable initial conditions are out of their specified bounds:
traj.phase0.controls:u
val: [-4.12229172e-01 -3.66856033e-01 -2.80844717e-01 -1.99433900e-01
-1.56629025e-01 -1.13824149e-01 -3.26813460e-02 3.99594324e-02
7.82792758e-02 1.16599119e-01 1.88859752e-01 2.54595136e-01
2.89272209e-01 3.22019211e-01 3.84095867e-01 4.40429629e-01
4.69970618e-01 4.99511606e-01 5.55510801e-01 6.01754139e-01
6.26148667e-01 6.50543194e-01 6.96432909e-01 7.36252629e-01
7.57258536e-01 7.76764323e-01 8.11593333e-01 8.44025167e-01
8.61133778e-01 8.74786177e-01 9.00666263e-01 9.24332967e-01
9.35771984e-01 9.43015357e-01 9.56746213e-01 9.67412856e-01
9.72945608e-01 9.78478361e-01 9.88966490e-01 9.92544685e-01
9.94432273e-01 9.96319861e-01 9.99711473e-01 9.99900358e-01
1.00000000e+00 1.00000000e+00 1.00000000e+00 1.00000000e+00
1.00000000e+00 1.00000000e+00 1.00000000e+00 1.00000000e+00
1.00000000e+00 1.00000000e+00 1.00000000e+00 9.99740247e-01
9.99450200e-01 9.99258930e-01 9.99176007e-01 9.99450671e-01
9.99971337e-01 1.00000000e+00 1.00000000e+00 1.00000000e+00
1.00000000e+00 9.94376352e-01 9.91409736e-01 9.88443120e-01
9.82634033e-01 9.73641857e-01 9.68898257e-01 9.61759824e-01
9.47868541e-01 9.33576031e-01 9.26036372e-01 9.17337810e-01
9.00848434e-01 8.83799004e-01 8.74540387e-01 8.65184313e-01
8.47077667e-01 8.28556952e-01 8.18786808e-01 8.08504624e-01
7.89013265e-01 7.69363561e-01 7.58923032e-01 7.43086617e-01
7.12795052e-01 6.82200510e-01 6.66061117e-01 6.49990808e-01
6.19527224e-01 5.89229486e-01 5.73325023e-01 5.57826517e-01
5.25226053e-01 4.83406684e-01 4.43294061e-01 4.13000503e-01
3.99078709e-01 3.85309120e-01 3.59732043e-01 3.34741315e-01
3.21558076e-01 3.09334330e-01 2.86162461e-01 2.63727435e-01
2.52240532e-01 2.30177275e-01 1.90980516e-01 1.55461203e-01
1.38245992e-01 1.30083184e-01 1.15162478e-01 1.00859286e-01
9.33139920e-02 8.66697400e-02 7.40746042e-02 6.21456861e-02
5.61676520e-02 5.05398132e-02 4.03412789e-02 3.06673327e-02
2.55640820e-02 2.12307448e-02 1.30162806e-02 5.29562823e-03
1.45610297e-03 -2.09307222e-03 -8.42703629e-03 -1.43210841e-02
-1.74303431e-02 -2.00137259e-02 -2.49108999e-02 -2.94053695e-02
-3.15860480e-02 -3.35550262e-02 -3.70454543e-02 -4.02656261e-02
-4.19643480e-02 -4.32572588e-02 -4.57081573e-02 -4.79219719e-02
-4.89777987e-02 -4.98945653e-02 -5.14441801e-02 -5.27836126e-02
-5.34901971e-02 -5.39821367e-02 -5.49146790e-02 -5.56740064e-02
-5.59927312e-02 -5.62163692e-02 -5.65527461e-02 -5.67913596e-02
-5.69172343e-02 -5.68963869e-02 -5.68568678e-02 -5.67317258e-02
-5.66252557e-02 -5.65340427e-02 -5.63315186e-02 -5.59258451e-02
-5.54461032e-02 -5.48282626e-02 -5.42864600e-02 -5.38344855e-02
-5.36309249e-02 -5.28657446e-02 -5.12836339e-02 -4.94748737e-02
-4.84485105e-02 -4.73832375e-02 -4.51796327e-02 -4.26966098e-02
-4.12894661e-02 -4.05585788e-02 -3.91000148e-02 -3.75598763e-02
-3.67474143e-02 -3.58012305e-02 -3.40076029e-02 -3.21440968e-02
-3.11280345e-02 -3.00611704e-02 -2.79053054e-02 -2.56004184e-02
-2.43845324e-02 -2.29623385e-02 -2.02663652e-02 -1.74457388e-02
-1.58988917e-02 -1.42704430e-02 -1.09826719e-02 -7.47069093e-03
-5.61803235e-03 -3.44135783e-03 6.84841995e-04 4.96599618e-03
7.29762507e-03 9.74935709e-03 1.46947968e-02 1.99727670e-02
2.27570294e-02 2.59487224e-02 3.19990359e-02 3.82615604e-02
4.16654645e-02 4.52108540e-02 5.22886756e-02 5.97651216e-02
6.37091356e-02 6.81692843e-02 7.66241381e-02 8.52583902e-02
8.98979372e-02 9.95291574e-02 1.19641680e-01 1.41581022e-01
1.53659193e-01 1.59820193e-01 1.71884153e-01 1.84377854e-01
1.90968597e-01 1.98131831e-01 2.11710772e-01 2.25219752e-01
2.32313024e-01 2.39659658e-01 2.54104885e-01 2.69129167e-01
2.77054854e-01 2.84920635e-01 2.99831353e-01 3.14893179e-01
3.22910066e-01 3.27717615e-01 3.38392026e-01 3.53279298e-01
3.70475877e-01 3.87061542e-01 4.01393104e-01 4.11423237e-01
4.15940615e-01 4.20767171e-01 4.29474812e-01 4.37689190e-01
4.42022481e-01 4.46340195e-01 4.54525044e-01 4.62547165e-01
4.66702151e-01 4.70092519e-01 4.75198877e-01 4.78830797e-01
4.80746726e-01 4.81492112e-01 4.82905098e-01 4.84366768e-01
4.85160839e-01 4.85352192e-01 4.85714929e-01 4.82748050e-01
4.81080619e-01 4.79413188e-01 4.76252333e-01 4.70641314e-01
4.67681360e-01 4.64721406e-01 4.59280678e-01 4.56763018e-01
4.55434889e-01]
lower: [-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75
-0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75 -0.75]
upper: [1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.
1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.
1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.
1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.
1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.
1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.
1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.
1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.
1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.
1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.
1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1. 1.
1. 1. 1. 1. 1. 1. 1. 1. 1.]
Set the initial value of the design variable to a valid value or set the driver option['invalid_desvar_behavior'] to 'ignore'.
Jacobian shape: (471, 679) (1.71% nonzero)
FWD solves: 0 REV solves: 30
Total colors vs. total size: 30 vs 471 (93.63% improvement)
Sparsity computed using tolerance: 1e-25.
Dense total jacobian for Problem 'problem7' was computed 3 times.
Time to compute sparsity: 0.8192 sec
Time to compute coloring: 0.4062 sec
Memory to compute coloring: 0.0000 MB
Coloring created on: 2026-02-26 12:24:25
Optimization terminated successfully (Exit mode 0)
Current function value: 5.358605984579545
Iterations: 6
Function evaluations: 7
Gradient evaluations: 6
Optimization Complete
-----------------------------------
==================================================
Grid Refinement - Iteration 4
--------------------------------------------------
Phase: traj.phases.phase0
Refinement Options:
Allow Refinement = True
Tolerance = 1e-06
Min Order = 3
Max Order = 14
Original Grid:
Number of Segments = 63
Segment Ends = [-1.0, -0.9778, -0.9556, -0.9333, -0.9111, -0.8889, -0.8667, -0.8444, -0.8222, -0.8074, -0.7926, -0.7778, -0.7333, -0.7185, -0.7037, -0.6889, -0.6667, -0.6444, -0.6222, -0.6, -0.5667, -0.5333, -0.4667, -0.4333, -0.4, -0.3333, -0.3, -0.2667, -0.2333, -0.2, -0.1667, -0.1333, -0.1, -0.0667, -0.0333, 0.0, 0.0333, 0.0667, 0.1333, 0.2, 0.2667, 0.3, 0.3333, 0.3667, 0.4, 0.4333, 0.4667, 0.5, 0.5333, 0.5667, 0.6, 0.6667, 0.7, 0.7333, 0.7667, 0.8, 0.8667, 0.8889, 0.9111, 0.9333, 0.9556, 0.9704, 0.9852, 1.0]
Segment Order = [5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 15, 5, 5, 5, 5, 5, 5, 5, 5, 5, 7, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 9, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 9, 5, 5, 5, 5, 5, 5, 5]
Error = [2.179e-05, 2.883e-06, 1.042e-05, 6.685e-06, 8.599e-06, 3.775e-06, 1.973e-06, 2.41e-06, 5.523e-07, 9.496e-07, 2.771e-06, 4.771e-08, 3.831e-07, 1.737e-06, 2.068e-06, 1.083e-06, 7.398e-07, 1.493e-08, 5.301e-07, 1.057e-06, 1.763e-06, 7.983e-06, 1.548e-06, 7.063e-06, 4.941e-07, 9.777e-07, 3.163e-06, 2.058e-06, 1.758e-06, 1.355e-06, 1.224e-06, 1.022e-06, 8.756e-07, 4.328e-07, 3.327e-07, 7.988e-08, 7.822e-07, 1.135e-06, 2.668e-06, 5.735e-07, 1.71e-06, 5.578e-07, 4.13e-07, 1.016e-06, 3.438e-07, 1.646e-06, 7.023e-07, 2.311e-06, 4.131e-06, 1.239e-05, 8.632e-06, 1.359e-05, 2.747e-06, 4.816e-06, 4.901e-06, 2.423e-06, 1.195e-06, 6.17e-08, 5.007e-06, 2.973e-06, 4.797e-07, 1.75e-06, 5.909e-06]
/home/runner/work/dymos/dymos/.openmdao-pixi/.pixi/envs/dev/lib/python3.13/site-packages/dymos/grid_refinement/hp_adaptive/hp_adaptive.py:379: RuntimeWarning: divide by zero encountered in divide
q_smooth = (np.log(self.error[phase_path][smooth_need_refine_idxs] /
/home/runner/work/dymos/dymos/.openmdao-pixi/.pixi/envs/dev/lib/python3.13/site-packages/dymos/grid_refinement/hp_adaptive/hp_adaptive.py:411: RuntimeWarning: divide by zero encountered in divide
q_split = np.log((self.error[phase_path][split_seg_idxs] /
/home/runner/work/dymos/dymos/.openmdao-pixi/.pixi/envs/dev/lib/python3.13/site-packages/openmdao/core/system.py:4656: OpenMDAOWarning:Calling `list_inputs` before `final_setup` will only display the default values of variables and will not show the result of any `set_val` calls.
/home/runner/work/dymos/dymos/.openmdao-pixi/.pixi/envs/dev/lib/python3.13/site-packages/dymos/phase/phase.py:3051: OpenMDAOWarning:phase0.states:x0 specifies 'fix_final=True'. If the given restart file has a different final value this will overwrite the user-specified value
/home/runner/work/dymos/dymos/.openmdao-pixi/.pixi/envs/dev/lib/python3.13/site-packages/dymos/phase/phase.py:3051: OpenMDAOWarning:phase0.states:x1 specifies 'fix_final=True'. If the given restart file has a different final value this will overwrite the user-specified value
Phase: traj.phases.phase0
New Grid:
Number of Segments = 93
Segment Ends = [-1.0, -0.9889, -0.9778, -0.9667, -0.9556, -0.9444, -0.9333, -0.9222, -0.9111, -0.9, -0.8889, -0.8778, -0.8667, -0.8556, -0.8444, -0.8333, -0.8222, -0.8074, -0.7926, -0.7852, -0.7778, -0.7556, -0.7333, -0.7185, -0.7037, -0.6889, -0.6667, -0.6444, -0.6222, -0.6, -0.5667, -0.55, -0.5333, -0.4667, -0.4333, -0.4167, -0.4, -0.3333, -0.3, -0.2833, -0.2667, -0.25, -0.2333, -0.2167, -0.2, -0.1667, -0.1333, -0.1, -0.0667, -0.0333, 0.0, 0.0333, 0.0667, 0.1333, 0.1667, 0.2, 0.2667, 0.2833, 0.3, 0.3333, 0.3667, 0.4, 0.4333, 0.4667, 0.5, 0.5167, 0.5333, 0.55, 0.5667, 0.5833, 0.6, 0.6333, 0.6667, 0.6833, 0.7, 0.7167, 0.7333, 0.75, 0.7667, 0.7833, 0.8, 0.8333, 0.8667, 0.8889, 0.9111, 0.9222, 0.9333, 0.9444, 0.9556, 0.9704, 0.9778, 0.9852, 0.9926, 1.0]
Segment Order = [5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 15, 15, 5, 11, 15, 7, 5, 5, 5, 7, 5, 5, 13, 13, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 11, 9, 7, 5, 5, 5, 5, 5, 13, 5, 5, 5, 5, 5, 5, 5, 7, 5, 15, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 9, 9, 9, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5]
Refined: True
Jacobian shape: (801, 1179) (1.21% nonzero)
FWD solves: 39 REV solves: 0
Total colors vs. total size: 39 vs 1179 (96.69% improvement)
Sparsity computed using tolerance: 1e-25.
Dense total jacobian for Problem 'problem7' was computed 3 times.
Time to compute sparsity: 1.7208 sec
Time to compute coloring: 0.8333 sec
Memory to compute coloring: -31.6836 MB
Coloring created on: 2026-02-26 12:24:29
Optimization terminated successfully (Exit mode 0)
Current function value: 5.3586078119379525
Iterations: 2
Function evaluations: 3
Gradient evaluations: 2
Optimization Complete
-----------------------------------
==================================================
Grid Refinement - Iteration 5
--------------------------------------------------
Phase: traj.phases.phase0
Refinement Options:
Allow Refinement = True
Tolerance = 1e-06
Min Order = 3
Max Order = 14
Original Grid:
Number of Segments = 93
Segment Ends = [-1.0, -0.9889, -0.9778, -0.9667, -0.9556, -0.9444, -0.9333, -0.9222, -0.9111, -0.9, -0.8889, -0.8778, -0.8667, -0.8556, -0.8444, -0.8333, -0.8222, -0.8074, -0.7926, -0.7852, -0.7778, -0.7556, -0.7333, -0.7185, -0.7037, -0.6889, -0.6667, -0.6444, -0.6222, -0.6, -0.5667, -0.55, -0.5333, -0.4667, -0.4333, -0.4167, -0.4, -0.3333, -0.3, -0.2833, -0.2667, -0.25, -0.2333, -0.2167, -0.2, -0.1667, -0.1333, -0.1, -0.0667, -0.0333, 0.0, 0.0333, 0.0667, 0.1333, 0.1667, 0.2, 0.2667, 0.2833, 0.3, 0.3333, 0.3667, 0.4, 0.4333, 0.4667, 0.5, 0.5167, 0.5333, 0.55, 0.5667, 0.5833, 0.6, 0.6333, 0.6667, 0.6833, 0.7, 0.7167, 0.7333, 0.75, 0.7667, 0.7833, 0.8, 0.8333, 0.8667, 0.8889, 0.9111, 0.9222, 0.9333, 0.9444, 0.9556, 0.9704, 0.9778, 0.9852, 0.9926, 1.0]
Segment Order = [5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 15, 15, 5, 11, 15, 7, 5, 5, 5, 7, 5, 5, 13, 13, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 11, 9, 7, 5, 5, 5, 5, 5, 13, 5, 5, 5, 5, 5, 5, 5, 7, 5, 15, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 9, 9, 9, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5]
Error = [1.877e-06, 5.981e-07, 4.368e-07, 4.098e-07, 5.77e-07, 1.505e-06, 9.251e-07, 7.015e-07, 1.745e-07, 3.864e-08, 8.728e-07, 7.71e-07, 9.694e-08, 5.498e-07, 4.723e-07, 6.638e-07, 6.397e-07, 1.092e-06, 2.544e-07, 3.533e-12, 4.493e-14, 4.37e-14, 3.908e-10, 7.251e-07, 7.463e-08, 7.411e-07, 5.303e-07, 2.007e-07, 3.889e-07, 2.355e-07, 1.183e-07, 1.037e-07, 3.647e-06, 3.346e-07, 8.659e-07, 5.231e-07, 3.081e-07, 3.495e-07, 6.587e-08, 1.488e-07, 2.53e-07, 1.507e-07, 1.974e-07, 2.519e-07, 7.71e-07, 1.239e-06, 1.281e-06, 6.485e-07, 3.454e-07, 3.157e-07, 2.118e-08, 6.712e-07, 5.424e-08, 1.816e-07, 2.991e-07, 3.297e-07, 1.019e-07, 1.794e-07, 7.232e-07, 5.058e-08, 3.155e-07, 1.146e-06, 9.937e-08, 4.574e-07, 7.22e-07, 4.197e-07, 1.411e-07, 1.138e-06, 2.465e-07, 2.504e-06, 3.022e-06, 2.204e-06, 2.857e-06, 5.713e-07, 1.012e-06, 7.521e-07, 5.568e-07, 4.664e-07, 3.859e-07, 2.595e-07, 9.897e-07, 2.148e-08, 2.368e-07, 7.178e-07, 3.576e-07, 3.45e-07, 3.164e-07, 8.848e-07, 1.45e-06, 2.359e-07, 2.949e-07, 9.72e-07, 3.496e-07]
Phase: traj.phases.phase0
New Grid:
Number of Segments = 102
Segment Ends = [-1.0, -0.9889, -0.9778, -0.9667, -0.9556, -0.9444, -0.9333, -0.9222, -0.9111, -0.9, -0.8889, -0.8778, -0.8667, -0.8556, -0.8444, -0.8333, -0.8222, -0.8074, -0.7926, -0.7852, -0.7778, -0.7667, -0.7556, -0.7444, -0.7333, -0.7185, -0.7037, -0.6963, -0.6889, -0.6667, -0.6444, -0.6222, -0.6, -0.5667, -0.55, -0.5333, -0.5, -0.4667, -0.4333, -0.4167, -0.4, -0.3333, -0.3, -0.2833, -0.2667, -0.25, -0.2333, -0.2167, -0.2, -0.1667, -0.1333, -0.1, -0.0667, -0.0333, 0.0, 0.0333, 0.0667, 0.1333, 0.1667, 0.2, 0.2667, 0.2833, 0.3, 0.3333, 0.3667, 0.4, 0.4333, 0.45, 0.4667, 0.5, 0.5167, 0.5333, 0.55, 0.5667, 0.5833, 0.5917, 0.6, 0.6167, 0.6333, 0.65, 0.6667, 0.675, 0.6833, 0.7, 0.7167, 0.7333, 0.75, 0.7667, 0.7833, 0.8, 0.8333, 0.8667, 0.8889, 0.9111, 0.9222, 0.9333, 0.9444, 0.9556, 0.9704, 0.9778, 0.9852, 0.9926, 1.0]
Segment Order = [11, 5, 5, 5, 5, 13, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 7, 5, 5, 15, 15, 15, 15, 5, 11, 15, 15, 7, 5, 5, 5, 7, 5, 5, 13, 13, 13, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 11, 15, 13, 5, 5, 5, 5, 5, 13, 5, 5, 5, 5, 5, 5, 5, 7, 9, 15, 15, 5, 5, 5, 5, 9, 5, 5, 5, 5, 5, 5, 5, 5, 5, 5, 7, 5, 5, 5, 5, 5, 9, 9, 9, 5, 5, 5, 5, 5, 13, 5, 5, 5, 5]
Refined: True
/home/runner/work/dymos/dymos/.openmdao-pixi/.pixi/envs/dev/lib/python3.13/site-packages/openmdao/core/system.py:4656: OpenMDAOWarning:Calling `list_inputs` before `final_setup` will only display the default values of variables and will not show the result of any `set_val` calls.
/home/runner/work/dymos/dymos/.openmdao-pixi/.pixi/envs/dev/lib/python3.13/site-packages/dymos/phase/phase.py:3051: OpenMDAOWarning:phase0.states:x0 specifies 'fix_final=True'. If the given restart file has a different final value this will overwrite the user-specified value
/home/runner/work/dymos/dymos/.openmdao-pixi/.pixi/envs/dev/lib/python3.13/site-packages/dymos/phase/phase.py:3051: OpenMDAOWarning:phase0.states:x1 specifies 'fix_final=True'. If the given restart file has a different final value this will overwrite the user-specified value
Jacobian shape: (1005, 1504) (1.11% nonzero)
FWD solves: 39 REV solves: 0
Total colors vs. total size: 39 vs 1504 (97.41% improvement)
Sparsity computed using tolerance: 1e-25.
Dense total jacobian for Problem 'problem7' was computed 3 times.
Time to compute sparsity: 2.4404 sec
Time to compute coloring: 1.1426 sec
Memory to compute coloring: -27.3633 MB
Coloring created on: 2026-02-26 12:24:34
Optimization terminated successfully (Exit mode 0)
Current function value: 5.358607995346617
Iterations: 1
Function evaluations: 2
Gradient evaluations: 1
Optimization Complete
-----------------------------------
Successfully completed grid refinement.
==================================================
Simulating trajectory traj
Done simulating trajectory traj
Problem: problem7
Driver: ScipyOptimizeDriver
success : True
iterations : 3
runtime : 4.5685E+00 s
model_evals : 4
model_time : 3.3843E-03 s
deriv_evals : 1
deriv_time : 3.8513E-02 s
exit_status : SUCCESS
(<Figure size 1000x800 with 5 Axes>,
array([<Axes: xlabel='time (s)', ylabel='$x_1$ (V)'>,
<Axes: xlabel='time (s)', ylabel='$x_0$ (V/s)'>,
<Axes: xlabel='time (s)', ylabel='J'>,
<Axes: xlabel='$x_0$ (V/s)', ylabel='$x_1$ (V)'>,
<Axes: xlabel='time (s)', ylabel='control u'>], dtype=object))
References#
Wikipedia contributors. Van der pol oscillator — Wikipedia, the free encyclopedia. 2020. [Online; accessed 12-June-2020]. URL: https://en.wikipedia.org/w/index.php?title=Van_der_Pol_oscillator&oldid=953857769.