TL;DR We built an optimization problem using OpenMDAO, Dymos, and the basic SIR model. The code is on github here.
The corona virus pandemic is being tracked with a set of mathematical models, that are tuned to match the data on the virus. They predict the number of deaths we’ll see and health officials are relying on them to track the “curve”, and to figure out when it will be safe for us to stop social distancing.
However, we’ve read a number of articles that clearly indicate that the modeling isn’t simple, or even widely agreed upon. One of the models being used is the Susceptible-Infected-Recovered (SIR) model. The SIR model is actually just an ODE with a few key parameters that can be tuned to match the data.
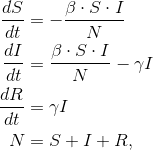
When we saw that, we thought that perhaps some of our optimization tools could be used to attack this problem. Dymos is a library we’ve written specifically to deal with time-integration and optimal-control problems. So Tristan Hearn, a member of the OpenMDAO Application team, has built a basic implementation and showed that it could be used to do optimization on defining the counter measures.
Now for a disclaimer
We are by no means infectious disease experts, but rather pracitioners of numerical optimization and multidisciplinary systems analysis. These models have not been tuned to match the data, and are by no means an accurate representation of the current situation anywhere in the world!
The Model
You can check out all the details on his pandemic repository. The code is set up to produce results like this:
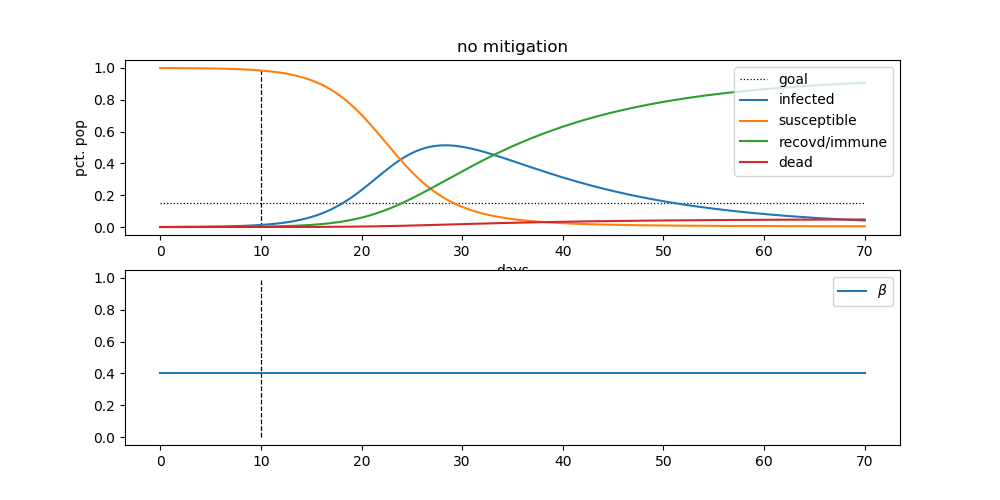
The blue line, represents the number of infected people. This is the “curve” that we’re all trying to flatten through social distancing. The goal is to lower the height of that hump, in order to not overwhelm our health care system.
By doing that, we can hopefully minimize the height of the red line. The red line represents the number of deaths from the pandemic. This is the scary one.
We don’t know if this model is useful to any actual infectious disease experts. Most probably, it won’t. But to be frank… it just feels good to be applying our skill sets to this world problem. If anyone out there wants to mess with the model, feel free!
Here are some things that need to be done:
- Parameter Identification: The model needs to be matched to real data. This isn’t easy. The data is different from state-to-state and county-to-county… this is part of what makes this so hard. See this fivethirtyeight article that talks about all of the uncertainty there is.
- Uncertainty quantification: The OpenMDAO team doesn’t have a lot of UQ experience, but we do have some of the most flexible and most scalable ODE optimization tools around. We suspect there are some good ways that UQ could be brought to bear here. If anyone wants to work on that, feel free to reach out.
- Model extension: Again, we’re not infectious disease experts. Those who are may be using more advanced models than this … or perhaps using a more complex version of this where different regions are modeled independently and interact with each other. We just don’t have the experience to know what to do here!
So if you’re looking for some interesting optimization problems to keep your busy during these strange times, check out Tristan’s pandemic repository.

